From serendipity to essential protac profiling
The microcosmos of biological processes is complex and full of surprises.
This is also the case for a new preprint from Richert, Nůsková et al. While developing rabeprazole-thalidomide hybrids as targeted protein degraders, the authors observed rapid, reversible ATP depletion in cells. Surprisingly, this effect was attributed to PROTAC molecules with a certain molecular structure.
𝗠𝗲𝗰𝗵𝗮𝗻𝗶𝘀𝗺
Mechanistic studies traced the effect to inhibition of mitochondrial complex I. Oxygen consumption was dose-dependently suppressed and restored by a complex II substrate, pinpointing mitochondrial complex I as the target.
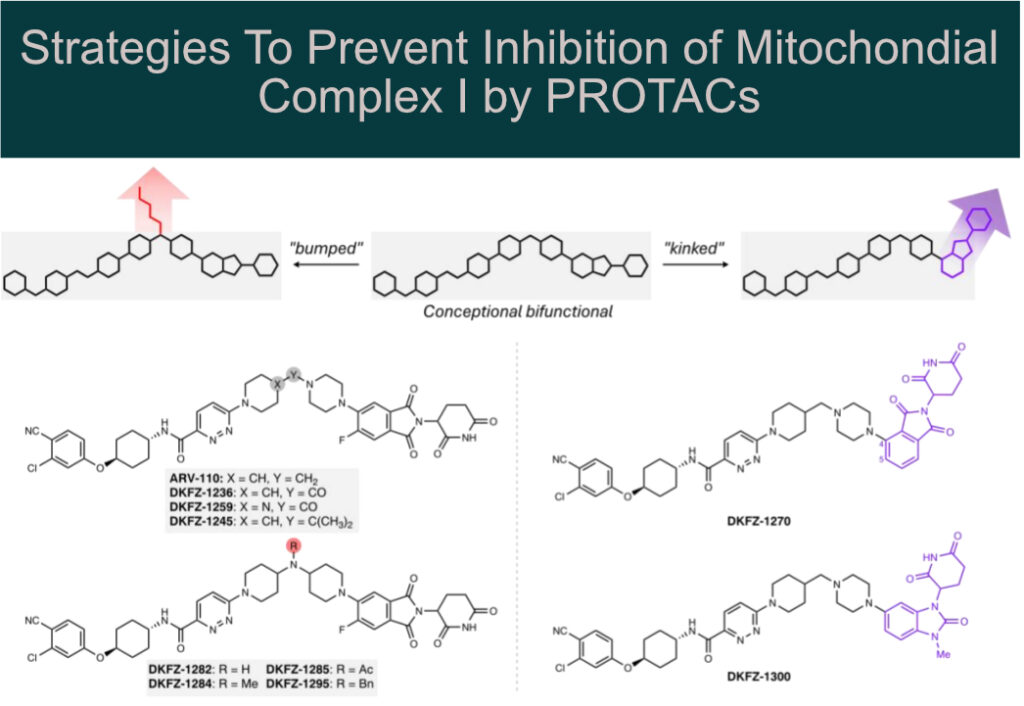
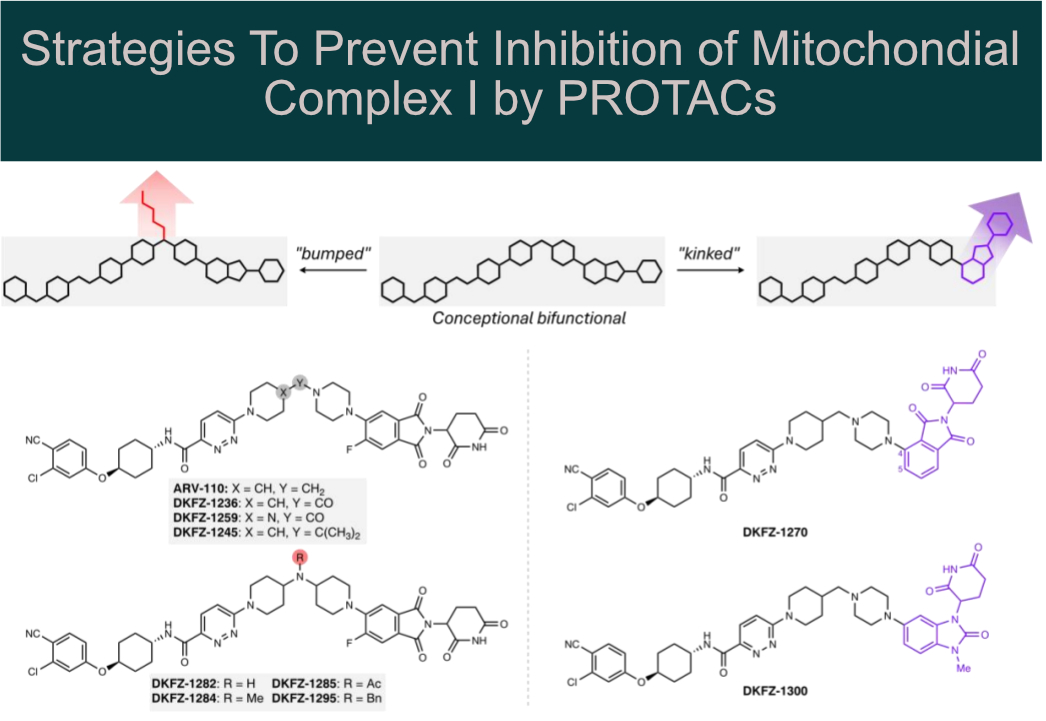
Systematic structural modification of PROTACs revealed that mitochondrial complex I inhibition occurs only in the presence of long, linear molecules that bind in the elongated (~30 Å) hydrophobic ubiquinone-binding tunnel.
𝗖𝗹𝗶𝗻𝗶𝗰𝗮𝗹 𝘀𝘁𝗮𝗴𝗲 𝗣𝗥𝗢𝗧𝗔𝗖𝘀 𝗮𝗹𝘀𝗼 𝗶𝗻𝗵𝗶𝗯𝗶𝘁 𝗺𝗶𝘁𝗼𝗰𝗵𝗼𝗻𝗱𝗿𝗶𝗮𝗹 𝗰𝗼𝗺𝗽𝗹𝗲𝘅 𝗜
The authors then screened 13 cereblon-recruiting PROTACs and found three clinical stage androgen receptor degraders to be potent inhibitors of mitochondrial complex I:
🔸 ARV-110: cell-free CI assay pIC50 = 7.66 (22 nM), nearly equipotent with rotenone
🔸 BMS-986365: CTG/2-DG pEC50 = 6.22 (0.60 µM)
🔸 ARV-766: submicromolar activity with partial inhibition
𝗦𝗼𝗹𝘂𝘁𝗶𝗼𝗻
Structural modification by introducing:
🔸 “bumps” (e.g., N-acylation of a piperidine linker nitrogen)
🔸 “kinks” (e.g., using a 1,3-substituted benzimidazolone cereblon recruiter)
A 384-well compatible CTG/2-DG assay was developed to enable routine CI liability screening.
This allowed the authors to abolish mitochondrial complex I inhibition while preserving AR degradation potency in cell-based assays.
𝗧𝗵𝗲 𝗸𝗲𝘆 𝘁𝗮𝗸𝗲𝗮𝘄𝗮𝘆
Mitochondrial complex I inhibition by PROTACs is not target- or recruiter-specific. It appears to be a general feature of long, linear, hydrophobic molecules, a common molecular architecture across bifunctional modalities. The authors propose rational geometric redesign as a generalizable mitigation strategy.
Are you going to implement this assay to your PROTAC development workflows? Let me know your thoughts!
Full preprint: https://lnkd.in/gWC2_jmk
Share your thoughts and leave your comment under my LinkedIn post here.